In
vitro
Transcription for RNA in situ Probes
Coordinator: Mike Shapiro
Purpose: To generate a labeled RNA molecule complementary
to an endogenous mRNA transcript of interest for use in in situ hybridization. In situ hybridization will allow us to visualize the
expression pattern of a gene in an organism.
I.
Linearize
plasmid. (This has already been done for you)
a. Choose an enzyme that will cut the plasmid on the
side opposite of the promoter (T3 or T7 in the case below) being used to
transcribe RNA.
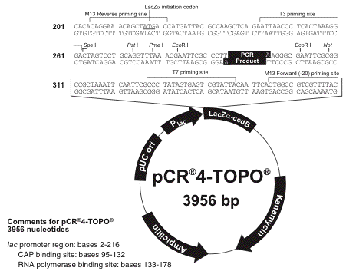
b. Choose the appropriate RNA polymerase to
transcribe your probe.
i. You will need to determine the orientation of
your sequence (ÒPCR ProductÓ in the above diagram) in the plasmid.
ii. RNA polymerases transcribe the compliment of the
sequence. If you are making an anti-sense
probe you will want to choose the polymerase whose promoter is 3Õ of your
sequence (when it is oriented so you could read the ORF). Use the polymerase on the 5Õ side if
you want to make a sense control probe.
Make sure you cut with an enzyme on the opposite side of the plasmid
insert as your desired RNA polymerase promoter.
c. Set up restriction digest reaction (25 µl
reaction)
12.5 µl purified DNA (or more up to 5 ug total)
2.5 µl 10X buffer
0.25 µl 100 mg/ml BSA (optional)
1 µl enzyme
add sterile MilliQ H2O up to 25 µl
d. Incubate at appropriate temperature for 2 hours
to overnight.
e. Stop reaction by incubating at 70 degrees for 20
minutes.
f. Check digestion by running 1 µl on 0.8% agarose,
1X TAE gel.
II.
Transcribe
RNA. (START HERE)
a. Set up the in vitro transcription reaction
5
µl digested DNA
4
µl 5X optimized transcription buffer (Promega #P1181)
2
µl 100mM DTT (Promega #P1171)
2 µl DIG RNA labeling mix (Roche #1277073)
0.5
µl RNasin (RNase inhibitor)
5.5
µl nuclease-free water
1 µl RNA polymerase, usually T3 or T7
b. Incubate at 37 degrees for 2 hours.
c. Remove 1 µl of reaction and dilute in 9 µl of
nuclease-free water in a new tube. Keep this sample on ice or in the freezer.
This sample will be run on a 1% agarose, 1X TAE gel, with a DNA or RNA ladder
and 2 µl of DIG-labeled control RNA (Roche # 1585746) to estimate yield. The
RNA bands should be approximately 10x brighter than the plasmid DNA.
d. Optional: Remove plasmid (DNA) template by DNAse
digestion
i. Add 2 µl DNAse I, RNase-free (Roche #0776785)
ii. Incubate at 37 degrees for 30 minutes
e. Precipitate RNA by adding
5 µl 0.1M EDTA pH8.0
1
µl 1mg/ml Glycogen (Ambion #9510)
3.1
µl 4M LiCl
90
µl EtOH
f. Mix and store at Ð80 degrees for 90 minutes to
overnight
g. Spin at 14,000 rpm at 4 degrees for 15 minutes.
h. Carefully remove supernatant and wash with 200 µl
70% EtOH.
i. Spin 2 minutes at 14,000 rpm at RT.
j. Remove supernatant. Spin again for 5 seconds and
carefully remove all traces of EtOH with a pipette.
k. Air dry pellet for 5 minutes.
III.
Hydrolyze
probe
a. Resuspend pellet in 50 µl of 40mM sodium
bicarbonate and 60mM sodium carbonate.
b. Heat probe for 20 minutes at 60 C.
c. Precipitate RNA by adding 200 µl nuclease-free
water, 25 µl 3M sodium acetate (NaOAc), 600 µl ethanol, and 1 µl glycogen to
each tube.
d. Incubate reaction at -20 C for at least 2 hours
(overnight is preferable).
e. Spin for 10 minutes at 14,000 rpm at 4 C.
f. The pellet may be very faint or even invisible at
this point. Carefully decant solution and add 500 µl 70% EtOH. Repeat one
additional time.
g. Air dry pellet for 5 minutes.
h. Resuspend pellet in 100 µl nuclease-free water.
i. Run 4 µl synthesized probe on 1% agarose, 1X TAE
gel with a DNA or RNA ladder. The hydrolyzed product should be between 200-300
bases.